The neurosphere assay: An effective technique to study neural stem cells
This video is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives (CC BY-NC-ND) license. You are free to share it for non-commercial purposes with proper attribution, but no modifications or adaptations are allowed.
1. Why use this method?
The neurosphere assay is a widely used technique to study neural stem cells (NSCs) because it allows for easy assessment of key properties like self-renewal, proliferation, and differentiation. It provides a simple in vitro model for evaluating NSC behavior, including their ability to form neurospheres and differentiate into neurons, astrocytes, and oligodendrocytes. The assay is also valuable for drug screening and toxicology studies. Its flexibility and reproducibility make it an effective tool for examining the effects of growth factors and signaling pathways on NSC biology, and it can be performed using both human and animal-derived NSCs.
2. What you’ll need
Materials
- 12 mm diameter coverslips
- 15 ml sterile falcon tubes
- 24 well plates
- 60 mm Petri Dishes
- BSA
- Calcium and Magnesium-free Hanks’ balanced Saline Solution
- Cell culture CO2 incubator
- Centrifuge
- Chemical dissociation kit (mouse)
- Curved Forceps
- Dissociation PBS preparation:
- KCl
- KH2PO4
- NaCl
- Na2HPO4
- EDTA
- Filter paper (pore size -11 µm)
- Hemocytometer
- Laminar flow
- Laminin (for coating)
- Microscope
- NeuroCult Chemical Dissociation Kit (Mouse)
- P1000 micropipette
- P1000 pipette tips
- Paraformaldeyhyde
- Poly- L – ornithine (for coating)
- Poly-D-lysine (for coating)
- RNAse free eppendorfs
- Scissor
- Serum Free Medium Preparation:
- DMEM/F12 Glutamax
- Penicillin / streptomycin
- B27
- EGF
- BFGF
- Spatula
- Tissue chopper
- Triton x-100
- Trypan Blue
- Trysin – EDTA
Antibodies to be used:
- Primary Antibodies:
- Anti-CD140a (PDGFRα) (rat)
- Anti-Chondroitin Sulphate Proteoglycan NG2 (rabbit)
- Anti-Doublecortin (rabbit)
- Anti-Doublecortin (chicken)
- Anti-Glial Fibrillary Acidic Protein (rabbit)
- Anti-Myelin Basic Protein (rabbit)
- Anti-Nestin (mouse)
- Anti-Neuronal Nuclei (mouse)
- Anti-SOX2 (rabbit)
- Anti-Tubulin β3 (rabbit)
- Secondary Antibodies:
- Alexa Fluor 488 donkey anti-chicken IgG (H+L)
- Alexa Fluor 488 donkey anti-rabbit IgG (H+L)
- Alexa Fluor 488 donkey anti-rat IgG (H+L)
- Alexa Fluor 568 donkey anti-mouse IgG (H+L)
- Alexa Fluor 568 donkey anti-rabbit IgG (H+L)
- Alexa Fluor 647 goat anti-mouse IgG (H+L)
- Anti-5-Bromo-2-Deoxyuridine
3. Step-by-step instructions
1. Basic setup and preparation of culture medium
1.1 Growth medium preparation – day of dissection
Depending in the number of pups used*, prepare serum-free medium (SFM) composed of Dulbecco’s modified eagle medium [(DMEM)/F12 with L-glutamine], supplemented with 100 U/mL penicillin and 100 µg/mL streptomycin (pen/strep), 1% B27, with also 10 ng/mL EGF and 5 ng/mL bFGF. Warm the culture medium to 37 °C in a water bath.
* for 5 pups prepare ~100 mL (50 mL for SVZ and 50 mL for DG); however, after counting the number of cells (step 5.1) the exact volume will have to be adjusted.
1.2 Microdissection of SVZ and DG
Preparation of 50−100 mL of calcium and magnesium-free Hanks’ balanced saline solution (HBSS) dissection medium, supplemented with 100 U/mL pen/strep.
1.3 Dissection Set up preparation
Prepare the tools needed for the dissection – microscope, scissor, and spatula, soak them in ethanol 70%.
2. Harvesting of postnatal (P1−3) mouse brains and SVZ/DG microdissections
2.1 Set up collection tubes and Petri dish
Fill a Petri dish (60 mm) with the solution prepared in 1.2 and 2 sample tubes with 500 uL of HBSS.
2.2 Euthanize Mice Pups
According to the protocol approved by the Institutional Animal Care facility/guidelines, euthanize mice pups (P1−3). For the decapitation, perform a single incision, with a scissor, at the base of the brainstem.
2.3 Expose skull
With a small pointed scissors, make a midline incision in the skin over the entire length of the head, thus revealing the surface of the skull.
2.4 Expose Brain
Using small, pointed scissors, held at a shallow angle to minimize brain damage, initiate a longitudinal incision at the base of the skull and meticulously follow the sagittal suture.
With curved forceps, peel the skull to the sides and expose the brain.
2.5 Brain Isolation
Slide under the base of the brain, with a small spatula, to cut cranial nerves and blood vessels for brain isolation. Transfer the brain into a Petri dish containing cold supplemented HBSS solution.
2.6 Brain Slicing Preparation
Transfer the Petri dish containing the brain to the dissecting microscope and carefully orient the brain on its dorsal surface for examination at low magnification.
2.7 Meninges Removal
Holding the brain by the cerebellum, remove the meninges with fine forceps. Rotate the brain to the ventral view and remove the remaining meninges
2.8 Brain Slicing
Cut, with forceps, the cerebellum and discard it. Set the brain onto the filter paper (pore size -11 µm), already placed in a tissue chopper, with curved-pointed forceps.
Cut the brain into coronal sections of 450 micrometers thick with a tissue chopper. Transfer the sections to a new Petri dish containing cold supplemented HBSS
2.9 Separation of SVZ
Under a dissecting microscope, sequentially separate the coronal brain slices in an anterior-to-posterior direction until reaching those containing the lateral ventricles. Using fine forceps, carefully dissect the thin layer of tissue surrounding the lateral ventricle walls – corresponding to the SVZ, excluding the striatal parenchyma and corpus callosum.
To isolate the SVZ, position the forceps tips at the lateral corners of each lateral ventricle: one under the corpus callosum and the other adjacent to the ventral area of the lateral ventricle. Then, make a small incision along the tissue surrounding the lateral ventricle.
Collect the dissected SVZ tissue in the sample tube containing supplemented HBSS solution – to maintain the accuracy of SVZ sample exclude SVZ in slices where the lateral ventricles and the hippocampal formation begin to appear.
2.10 Separation of DG
Continue examining all remaining slices in an anterior-to-posterior direction until reaching the hippocampal formation. Using forceps, discard the first slice containing the hippocampus where the DG is not yet clearly distinguishable.
Isolate the hippocampus from the remaining slices. Refocus the microscope to precisely identify the borders of the DG within the hippocampal structure.
Separate DG by cutting between DG and CA1 region, followed by a vertical cut between DG and CA3 region, with forceps. Discard fimbria and any adjacent tissue.
Collect the isolated DG into a sample tube containing supplemented HBSS solution.
3. Tissue dissociation
3.1 Dissociation of SVZ and DG
To dissociate the SVZ and DG tissue present in their respective tubes, add trypsin-EDTA 0.05%. The final concentration of Trypsin-EDTA should be between 5% and 10% in HBSS. Incubate the samples at 37°C for approximately 15 minutes, until the tissue clumps together.
3.2 Wash Trypsin – EDTA
Perform four consecutive washes to remove the trypsin. Each wash involves aspirating the media and replacing it with 1 mL of fresh HBSS supplemented solution.
Remove the HBSS and resuspend the digested tissue in 1 mL of SFM supplemented with 10 ng/mL EGF and 5 ng/mL bFGF. Gently pipette the cell pellet up and down – mechanical dissociation – approximately 7-10 times using a P1000 pipette until a homogeneous cell solution is formed.
4. Cell-pair assay to study cell fate
- Prepare coated 24-well plates for adherent monolayer cultures according to sections 8-10.
- To count the number of SVZ or DG cells (obtained in section 3) to be plated, use a solution containing 0.2% Trypan blue and count the cells using a hematocytometer.
- Dilute the dissociated cell suspension in SFM supplemented with 5 ng/mL EGF and 2.5 ng/mL bFGF (low EGF/bFGF) at a density of 11,300 cells/cm2 and plate them on coated glass coverslips.
- After 24 h, fix the cells for immunocytochemistry against NSC markers. See section 14 for an immunostaining protocol of neuroshperes.
5. Expansion of postnatal neural stem cells as neurospheres
- To determine the density of the dissociated SVZ or DG cell suspension (obtained in section 3), count the cells using a hematocytometer.
- Dilute SVZ and DG single cell suspension at a density of 2 x 104 cells/mL in SFM supplemented with 10 ng/mL EGF and 5 ng/mL bFGF. Seed SVZ and DG cells in uncoated 60 mm Petri dishes with a final volume of 5 mL/Petri dish.
- Incubate SVZ and DG cells for 6−8 days and 10−12 days, respectively to form primary neurospheres, at 37 °C with 5% CO2.
- When the majority of neurospheres have a diameter of 150−200 µm, perform the neurosphere passage.
6. Passaging of neurospheres
- To passage neurospheres, collect the SFM with growth factors containing neurospheres from the 60 mm Petri dish(es) and centrifuge for 5 min at 300 x g.
- Discard the supernatant and resuspend the neurosphere pellet using a chemical dissociation kit (mouse) according to the manufacturer’s instructions (Table of Materials).
- Centrifuge for 5 min at 300 x g, remove the supernatant and add 1 mL of SFM supplemented with 10 ng/mL EGF and 5 ng/mL bFGF.
- Triturate up and down approximately 10x with a P1000 pipette to dissociate neurospheres.
- Count the number of cells using a solution containing 0.2% Trypan blue and a hematocytometer.
- Reseed cells at a density of 2 x 104 cells/mL in uncoated 60 mm Petri dishes.
- Incubate SVZ and DG cells for 6−8 days and 10−12 days, respectively to obtain secondary neurospheres, at 37 °C with 5% CO2.
7. Storage of neurospheres for gene expression assay
- Collect the medium containing neurospheres (obtained from steps 5.3 and 6.7) from the 60 mm Petri dishes.
- Centrifuge for 5 min at 300 x g and discard the supernatant.
- Wash the cells 2x with 1 mL of HBSS (centrifuge 5 min at 300 x g between washes).
- Centrifuge for 5 min at 300 x g, discard the supernatant and store the pellet of neurospheres at -20 °C for molecular biology analysis.
8. PDL coating plate procedure
- To prepare solution 1 (0.1 M borate buffer), weigh 3.92 g of boric acid and dilute in 400 mL of high purity water. Adjust the pH to 8.2 and make up to 500 mL with high purity water.
- To prepare solution 2 (0.167 M borate buffer), weigh 10.3 g of boric acid and dilute in 900 mL of high purity water. Adjust the pH to 8.2 and make up to 1,000 mL with high purity water.
- To reconstitute poli-D-lysine (PDL) (1 mg/mL in 0.1 M borate buffer), dilute 100 mg of PDL in 100 mL of solution 1.
- Make aliquots of 10 mL to use immediately or freeze and store at -20 °C.
- Under the laminar flow, add 1 coverslip per well and sterilize under UV light for 15 min.
- Use the reconstituted PDL or thaw frozen reconstituted PDL.
- Prepare the final solution of 100 µg/mL PDL in 0.167 M borate buffer by adding 10 mL of reconstituted PDL to 90 mL of solution 2.
- Add the final solution to wells for a minimum of 2 h to overnight at 37 °C.
NOTE: For 24-well plates, add a volume of 500 µL in each well. - Remove the solution and wash 3x with high purity water.
- Let the coverslips dry in the laminar flow hood.
- Leave the multi-well culture plates at 4 °C.
9. PDL/Laminin coating plate procedure
- On day 1, coat the plates with PDL as described in section 8.
- On day 2, remove the PDL solution and wash 3x with high purity water. Let dry.
- Prepare 5 µg/mL laminin in cold SFM devoid of growth factors.
- Add dissolved laminin to the coverslips and incubate at 37 °C overnight.
NOTE: For 24-well plates, add a volume of 500 µL in each well. - Remove laminin using a pipette. NOTE: Do not wash the coverslips from laminin.
- Use immediately or store at -20 °C.
10. Poly-L-ornithine (PLO) /laminin coating procedure
- Under the laminar flow, add one coverslip per well and sterilize under UV light for 15 min.
- Add 0.01% PLO solution to each well for 20 min at room temperature (RT).
NOTE: For 24-well plates, add a volume of 500 µL in each well. - Remove the solution and wash 3x with sterilized 1x PBS. Let dry.
- Prepare 5 µg/mL laminin in sterile 1x PBS.
- Incubate for 2 h at 37 °C.
- Remove laminin. NOTE: Do not wash the coverslips from laminin.
- Use immediately. NOTE: Make sure that the coverslip is fully covered by the PLO solution by gently tapping the coverslip with a pipette tip. When shaken, the multi-well plates should make no sound.
11. Evaluation of neuritogenesis by generating a differentiated monolayer of cell
- Collect media containing neurospheres from 60 mm Petri dishes (obtained from section 5) and centrifuge for 5 min at 300 x g at RT.
- Discard the supernatant and dissociate the pellet of neurospheres in 1 mL of dissociation PBS (i.e., PBS without Mg2+/Ca2+ and with EDTA [2.7 mM KCl, 1.5 mM KH2PO4, 137 mM NaCl, 8.1 mM Na2HPO4 and 0.5 mM EDTA 4Na, at pH = 7.40]) by incubating for 15 min followed by mechanical dissociation. Alternatively, dissociate neurospheres using a chemical dissociation kit (mouse) (Table of Materials).
- Centrifuge for 5 min at 300 x g at RT and discard the supernatant.
- Resuspend the cell pellet in 1 mL of SFM devoid of growth factors.
- Determine cell density using a hematocytometer.
- Dilute the dissociated cell suspension in SFM devoid of growth factors at a density of 3,766 cells/cm2 and plate cells on coated glass coverslips in 24-well plates.
- After 1−3 days, fix cells for immunocytochemistry against a protein of the cytoskeleton (see section 14).
12. Differentiation of neurosphere cultures
Neurospheres obtained from cell expansion, either from primary or passaged neurospheres (obtained in sections 5 or 6) can be differentiated into cells from different neural lineages.
12.1 Neurosphere collection
When neurospheres have a diameter of 150−200 µm, collect 25 µL of neurosphere suspension medium and plate on coated glass coverslips, in 24-well plates.
Note: Cell survival, proliferation and differentiation can be analyzed using different cell assays.
13.Cell biology assay
13.1 Self-renewal capacity
- Seed SVZ and DG cells at a density of 1.0 x 104 cells/mL (in uncoated 24-well plates) in growth SFM medium containing 5 ng/mL EGF and 2.5 ng/mL bFGF (low EGF/bFGF).
- Count the number of resulting primary and secondary neurospheres.
13.2 Cell survival assay
Expose plated neurospheres to 3 µg/mL propidium iodide (PI) for 30 min before cell fixation in the incubator at 37 °C.
Note: PI is an autofluorescent agent that is only able to enter cells with compromised membrane integrity. Other methods to analyse cell survival can be used such as caspase 3 staining or the terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay.
13.3 Cell proliferation assay
Expose plated neurospheres to 10 µM 5-bromo-2′-deoxyuridine (BrdU) for 4 h before fixation in the incubator at 37 °C.
13.4 Cell differentiation assay
- Expose 7-day-old plated neurospheres to 10 µM BrdU in the first 24 h, in the incubator at 37 °C.
- Renew the SFM devoid of growth factors (differentiative conditions) and allow cells to develop in the absence of BrdU for the following 6 days until fixation.
NOTE: These pulse-chase experiments, by co-labelling with markers of mature neural cells, allow the evaluation of progenitor cells that differentiate into mature cells during the protocol.
14. Immunostaining and neurosphere culture
14.1 Cell Fixation
- Prepare a solution of 4% paraformaldehyde (PFA) in 1x phosphate-buffered saline (PBS).
- Treat the cells by adding 500 µL of the prepared PFA solution to each well containing the cells. Allow the cells to be exposed to PFA for 20 minutes at room temperature (RT).
- After the fixation step, wash the cells three times with 1x PBS. Each wash should last for about 5 minutes
- Once washed, store the coverslips containing the fixed cells in 1x PBS at 4°C until further use.
14.2 Denaturation Method
- Prepare a 1 M HCl solution and heat it to 37°C.
- Rinse the coverslips containing the fixed cells three times with 1x PBS to remove any residual PFA.
- Permeabilize the cells by incubating them in a solution containing 1% nonionic surfactant (e.g., Triton X-100) in PBS for 30 minutes.
- Denature the DNA by treating the cells with the pre-heated 1 M HCl for 30-40 minutes at 37°C.
- After denaturation, wash the cells four times with 1x PBS to neutralize the acidic environment and remove any residual HCl.
14.3 Blocking
- Rinse the coverslips containing the fixed and, if applicable, denatured cells with 1x PBS for 5 minutes
- Incubate the coverslips with a solution containing 0.5% nonionic surfactant (e.g., Triton X-100) and 3% bovine serum albumin (BSA) in 1x PBS for 1.5 hours.
15. Incubation
- On the first day, without rinsing, expose the cells to primary antibodies diluted in a solution containing 0.1% nonionic surfactant and 0.3% BSA in 1x PBS. Incubate the coverslips overnight at 4°C, shielded from light if the antibodies are fluorophore-conjugated.
- On the second day, return the coverslips to their respective wells and wash them three times with 1x PBS for 5 minutes each to remove excess primary antibodies.
- Apply fluorescence-conjugated secondary antibodies (diluted 1:200) and Hoechst 33342 (at a concentration of 12 µg/mL) in 1x PBS to the coverslips. Incubate them for 2 hours at room temperature, protected from light in the incubation chamber.
- Wash the coverslips three times with 1x PBS for 5 minutes each to remove unbound secondary antibodies and Hoechst dye.
- Mount the coverslips onto microscope slides using fluorescence mounting medium, applying 5 µL of mounting medium per coverslip.
- Allow the coverslips to air dry at room temperature, shielded from light, for one day before microscopy.
- Observe and capture images using a fluorescent microscope.
4. Practical tips
Importance of each solution/reagent:
SFM – allows reproducibility and better control over physiological responsiveness. Serum-free media also gives you more precise control of what components your cell culture media has. This way, we can control experimental conditions more precisely.
DMEM/F12 – basal medium provides basic nutrients for the growth of various mammalian cell lines.
L-glutamine – energy source, since it is an essential amino acid.
Penicillin and streptomycin – antibiotic mixture to reduce contamination caused by gram+ and gram- bacteria.
B27 – a mixture of antioxidant enzymes, proteins, vitamins, and fatty acids to support neuronal survival in culture.
EGF and bFGF – promote the proliferation and differentiation of neural stem cells.
Hanks’ Balanced Salt Solution (HBSS) – to rinse chelators from the culture before cell dissociation.
For brain isolation: Using cold-supplemented HBSS solution – helps maintain the brain’s physiological state and minimize tissue degradation.
For tissue dissociation: Trypsin, a protease, effectively breaks down the proteins that hold cells together within the SVZ and DG tissue samples, using a low concentration that allows a controlled dissociation.
For meninges Removal: Removing the meninges provides a clear view of the brain structures and facilitates further dissection/slicing.
For cell counting: Trypan Blue is used as a vital stain to selectively color dead tissues or cells blue. Live cells or tissues with intact cell membranes are not colored.

It should be ensured that the seeding takes into account the number of live cells and not the total number of cells.
For gene expression: It is very important that all HBSS is removed from the pellet to prevent any influence on later cell lysis. Dry cell pellets can be stored indefinitely at -20ºC for up to 1 month at 4ºC. It should be remembered that cells are not viable for culture afterwards.
For the PDL coating plate: This coating serves as a substrate for cell adhesion and growth in cell culture experiments. Poly-D-lysine is a synthetic polymer composed of the amino acid lysine. When applied to the surface of culture plates, PDL forms a positively charged layer that promotes the attachment of cells. This is particularly useful when working with cells that require a positively charged surface for optimal adhesion, such as neurons or certain types of stem cells.
The buffer solution will be used to dissolve the PDL and pH need to be adjusted.
Another borate buffer solution (with a higher concentration) is needed in order to have two different concentrations of borate buffer that allows for flexibility in adjusting the pH and concentration of the PDL solution.
Sterilizing the plates or coverslips under UV light serves to eliminate any potential microbial contamination on the surfaces before they are used for cell culture experiments.
After incubating the plates to allow for effective coating, any excess PDL solution is removed by washing the wells with high-purity water. This helps to remove any unbound PDL and ensures that only the coated surface remains.
Drying the plates in the laminar flow hood ensures that the coating is properly set before proceeding with cell culture experiments. Once dry, the coated plates are stored at 4 °C until they are ready to be used for cell culture experiments. Storing the plates in the refrigerator helps to preserve the integrity of the PDL coating and ensures that the plates remain suitable for cell culture over an extended period.
For the laminin coating plate: Laminin is a protein found in the extracellular matrix that promotes cell adhesion and growth. It is important not to wash the coverslips or wells after removing laminin to ensure that the laminin-coated surface remains intact.
For Poly-L-Ornithine (PLO): PLO is a positively charged polymer that promotes cell adhesion. By coating the wells with PLO, an adhesive substrate is created for cells to attach to. During the 2h incubation, laminin binds to the PLO-coated surface, providing an optimal substrate for cell adhesion and growth.
For the generation of a differentiated monolayer of cells: It is necessary to dissociate the neurospheres into single cells before plating them onto coverslips.
Determining cell density using a hematocytometer, ensures that an appropriate number of cells are plated onto coverslips for subsequent experiments. This cell density is optimized for promoting cell adhesion and growth while avoiding overcrowding on the coverslips.
For cell survival assay: Live cells have intact membranes, since PI can only enter cells with compromised membrane integrity – dead cells – this dye allows to assess cell survival.
For cell proliferation assay: BrdU will be incorporated in the new synthetized DNA of active proliferative cells.
For cell Fixation: Fixation preserves the cellular structure and allows for the visualization of specific proteins of interest using immunocytochemistry techniques. PFA is a commonly used fixative that helps preserve cellular structures. It is important to keep this solution cold to maintain its stability, either at 4°C or -20°C. This treatment helps to immobilize the cells and prevent any further changes or degradation. It is crucial to have a washing step with PBS for removing excess PFA and any debris from the cells while ensuring their integrity.
After Fixation cells, can be stored at 4ºC in PBS1X., but they should not be allowed to dry out.
For denaturation: Usage of 37ºC HCl will help in denature DNA by breaking DNA strands and expose the BrdU molecules previously incorporated during cell proliferation.
For cell Permeabilization: Usage of nonionic detergents such as TRITONX-100 helps to make the cell membrane more permeable, allowing subsequent reagents to penetrate the cells effectively.
For blocking prior to immunocytochemistry: This step permeabilizes the cells further and blocks nonspecific binding sites, ensuring specific antibody binding during the subsequent steps. It should be noted that if NeuN staining is desired, a higher concentration of BSA (6%) should be used.
5. Critical appraisal & implications for future research
The neurosphere assay is a valuable tool for studying neural stem cells (NSCs), but it has limitations, such as not fully replicating the in vivo environment and lacking interactions with other cell types. Despite this, it remains crucial for research on NSC self-renewal, differentiation, and disease modeling. Future improvements could include more realistic 3D cultures and co-cultures, as well as the use of advanced techniques like single-cell RNA sequencing and live imaging to deepen our understanding of NSC behavior. Enhancing scalability for high-throughput screening and regenerative medicine applications also holds promise for future research.
This protocol is part of the Braining project, co-founded by the European Union under the project “Collaborative learning and innovative teaching in brain drug screening” (2023-1-PL01-KA220-HED-000160284). It is licensed under a Creative Commons Attribution-NonCommercial (CC BY-NC) license, allowing sharing and adaptation for non-commercial purposes with proper attribution.


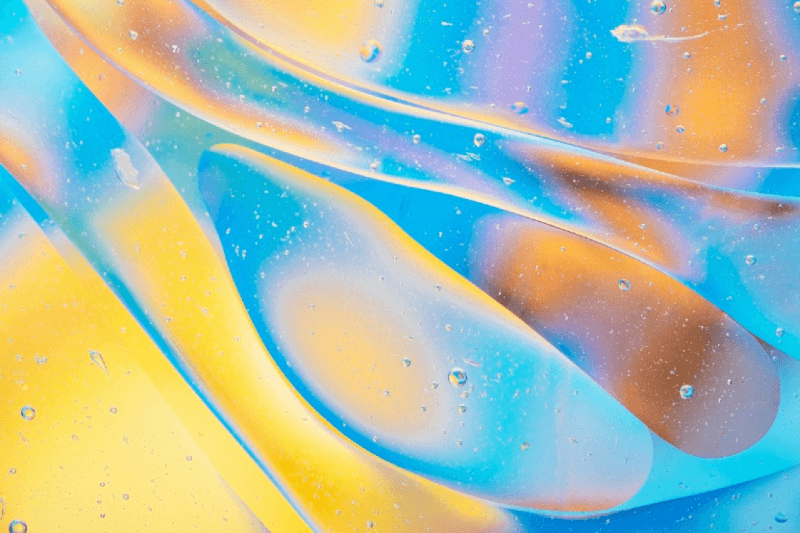
Adding comments is only possible for registered users.
Sign in